Genomics/Epigenomics
Session: Genomics/Epigenomics
369 - Identification of Genetic Disorders based on Phenotype and Subsequent Medical Management
Monday, May 6, 2024
9:30 AM - 11:30 AM ET
Poster Number: 369
Publication Number: 369.2749
Publication Number: 369.2749

Sara Strandlund, DO (she/her/hers)
Resident
Children's Mercy Hospitals and Clinics
Kansas City, Missouri, United States
Presenting Author(s)
Background: Many genetic disorders manifest in the neonatal period and contribute to significant morbidity and mortality. These patients often require significant support in a NICU while awaiting diagnosis. Symptom driven exome sequencing has emerged as a cost effective and relatively rapid means of identifying single gene disorders. This tool has been shown to help guide management of critically ill infants with suspected underlying genetic disorders.
Objective: The aim of this study is to evaluate the indications for genetic testing based on phenotype and compare these with the diagnostic yield of symptom driven exome sequencing (ES) and chromosome microarray in the NICU.
Design/Methods: A retrospective chart review was conducted of neonates admitted to the Children’s Mercy Kansas City level IV NICU and underwent genetic testing (included ES -rapid and non-rapid, microarray, gene panel and FISH) between January 1, 2022, to December 31, 2022. Neonates with well-defined chromosomal anomalies (Trisomy 13, 18, 21) were excluded to focus on single gene disorders. Medical management changes after results ranged from changes to medication, procedure, diet, new subspecialty involvement and recommended familial testing. Chi-square and Fisher’s exact tests were used to evaluate relationships between the phenotypes and the diagnostic yield of ES.
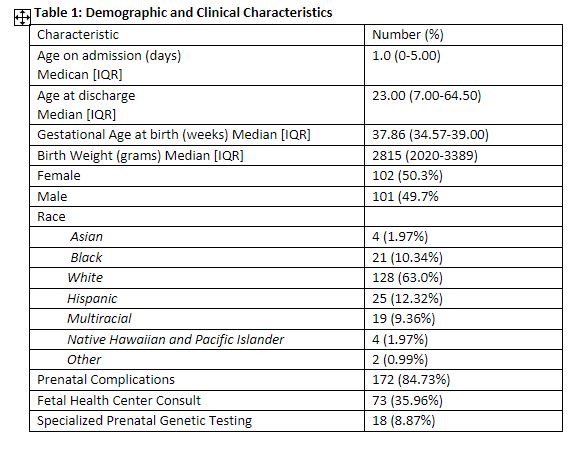
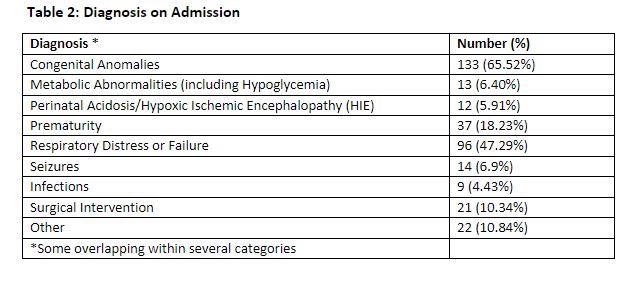
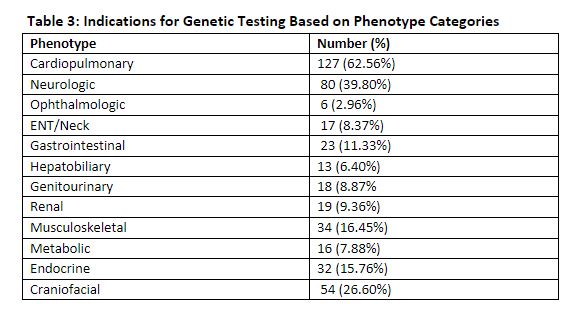
Results: 203 neonates underwent genetic testing. The median gestational age of patients was 37.8 weeks [IQR: 23.57-39.0] (Table 1). Most common admission diagnosis was congenital anomalies (65.5%) [ Table 2]. Cardiopulmonary phenotype (65.6%) was the most common indication for genetic testing (Table 3). 130/203 (64%) neonates underwent ES. 25/130 single gene disorders were identified with a diagnostic yield of 19.2%. Multiple factors account for the lower yield of ES in our study than previously reported. Our study was limited to 1 year with a smaller sample size. Neonates with cardiopulmonary concerns had a significant lower diagnostic yield of ES compared to other phenotypes (9% vs. 34.6%, p< 0.001), impacting the overall yield of testing. Management changes occurred in 48/203 (23.6%). For some neonates, no changes were recommended because appropriate management had been adjusted at the time of consultation.
Conclusion(s): This study highlights that genetic testing remains as an important adjunct to clinical care but not the only determining factor in care and management of critically ill neonates. Exome sequencing is well known to have diagnostic limitations, and this study demonstrates the importance of continued development of new technology for diagnosis of genetic disease.