Neonatology
Session: Neonatal Infectious Diseases/Immunology 1
593 - Metagenomic next-sequencing Based Development and Internal Validation of an Accurate Calculator for Diagnosis of Neonatal Sepsis
Friday, May 3, 2024
5:15 PM - 7:15 PM ET
Poster Number: 593
Publication Number: 593.616
Publication Number: 593.616

Meng Meng Ge
pediatrician
Children's Hospital of Fudan University
Shanghai, Shanghai, China (People's Republic)
Presenting Author(s)
Background: Neonatal sepsis is the leading cause of infectious disease-related deaths worldwide in children younger than 5 years yet remain challenging to diagnosis because of limitations in existing microbiologic tests. Prompt diagnosis and treatment of neonatal sepsis are crucial to prevent severe morbidity and mortality. However, the initial, clinical presentation is often subtle and nonspecific, and commonly used biomarkers have low predictive values for sepsis, which also presents a daily challenge to clinicians involved in neonatal care. mNGS methodologies are complex, multistep processes that currently lack standardization, further complicating result interpretation.
Objective: To develop and test a diagnosis calculator that can be used to assist clinicians to better interpret and apply mNGS.
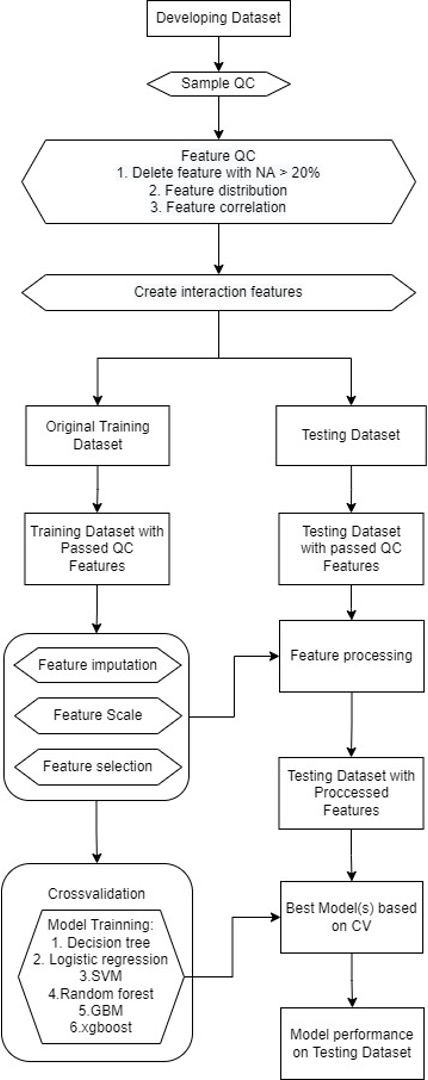
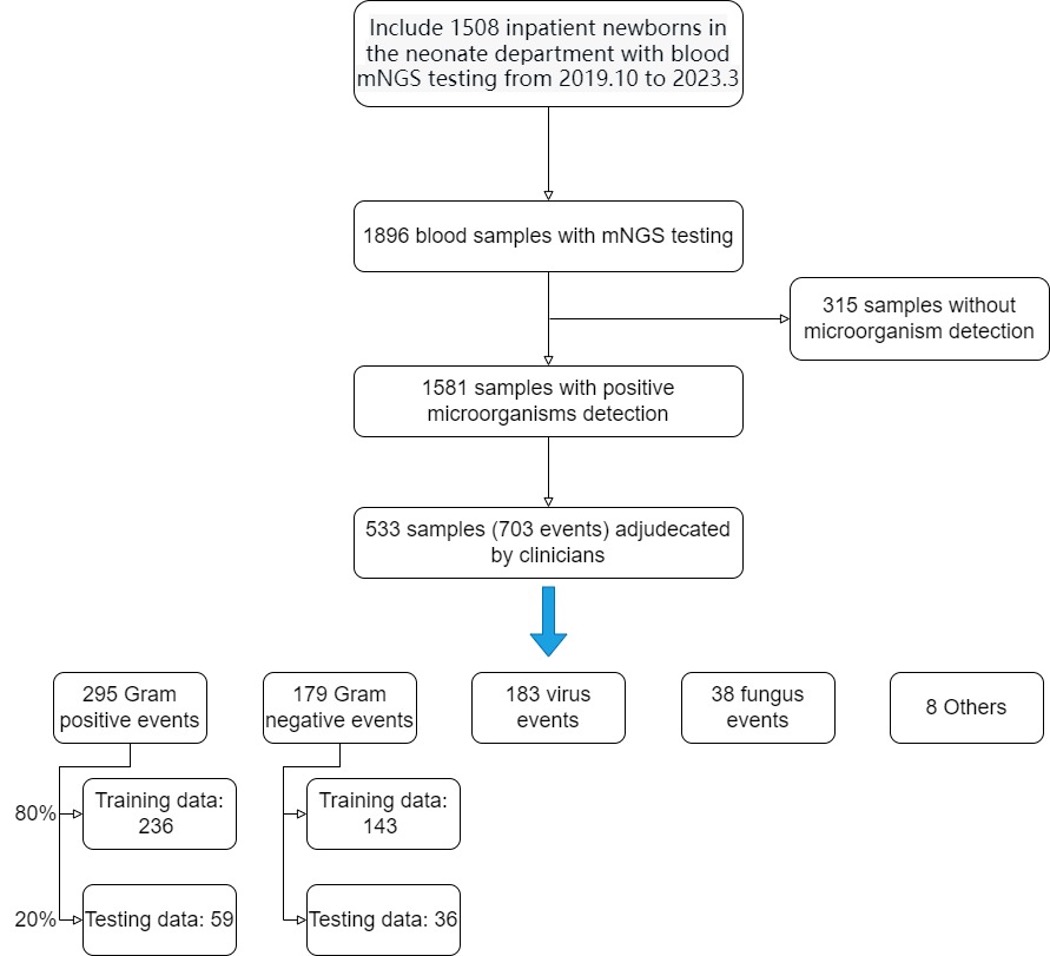
Design/Methods: The study cohort includes 1,508 inpatient newborns in the department of neonates with blood mNGS testing from January 1, 2019, through March 31, 2023. And, these are 1,896 blood samples with mNGS testing and 1,581 samples with positive microorganisms detection. Two multivariable lagistic regression models for predicting the potential pathogen of neonatal sepsis were trained and tested. With the use of blood/CSF culture-confirmed sepsis as the main outcome. A final diagnostic category was assigned for each patient, after review by at least two experienced neonatologists of all clinical, laboratory and imaging data.
Results: A total of 533 samples (703 events) adjudecated by clinicians, were included in the study. The gram-positive bacteria model included 295 events(training data included 236 events, testing data included 59 events), and the gram-negative bacteria model included 179 events(training data included 143 events, testing data included 36 events). These two models included four variables respectively. The G+ model revealed areas under the curve of 0.85 and recall value of 0.75, compared to manual diagnosis(AUC of 0.81, recall value of 0.58). The G- model revealed areas under the curve of 0.76 and recall value of 0.86, compared to manual diagnosis (AUC of 0.85, recall value of 0.81).
Conclusion(s): Establishing reliable predictive models will enable clinicians to more accurately interpret mNGS findings and may allow for more targeted early antibiotic therapies for those at highest risk of infection.