Infectious Diseases
Session: Infectious Diseases 3
126 - Genotypic and phenotypic characterization of pathogenic neonatal Escherichia coli strains
Sunday, May 5, 2024
3:30 PM - 6:00 PM ET
Poster Number: 126
Publication Number: 126.1703
Publication Number: 126.1703

Dustin D. Flannery, DO, MSCE
Assistant Professor of Pediatrics
Children's Hospital of Philadelphia
Philadelphia, Pennsylvania, United States
Presenting Author(s)
Background: There are no specific preventative measures for invasive neonatal E. coli infection. A better understanding of the current prevalence of specific clones, virulence factors, and resistance genes in E. coli infections among term and preterm infants may lead to improved screening, preventive interventions, and anti-virulence antibiotic-sparing therapies or vaccine targets.
Objective: To perform genetic and phenotypic characterization of pathogenic neonatal E. coli strains.
Design/Methods: Infants admitted to two academic NICUs were screened for invasive E. coli infection (blood, urine, CSF, or respiratory culture) from 2021-2023. Isolates were collected before discard and suspended in growth media supplemented with 15% glycerol. Isolates were sent to the Penn State E. coli Reference Center for whole genome sequencing and multiple locus sequence typing. Clinical data including lab-reported susceptibilities were abstracted from the medical record.
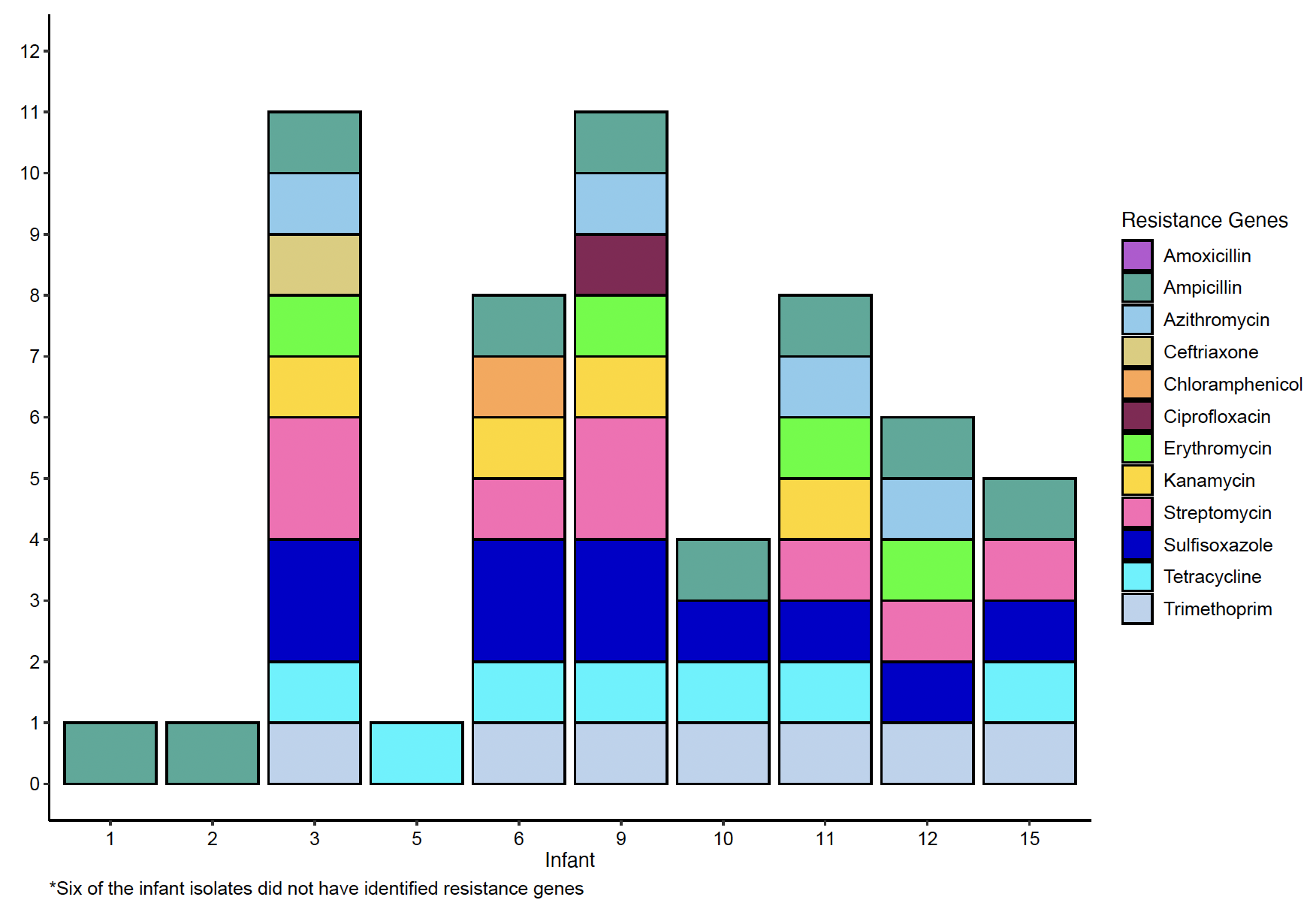
Results: Sixteen infants had E. coli infection (12 blood, 2 urine, 2 respiratory, no CSF). Median GA was 27.9 weeks, 12 (75%) were male, 7 (44%) were born via C-section, and 6 (38%) were transferred before discharge; none died before discharge (Table). Six infants had early-onset (first 3 days) and 10 had late-onset (after 3 days) infection. Mothers of all 16 infants received antibiotics during delivery admission, and 3 (19%) mothers had E. coli infection around delivery (1 blood, 1 urine, 1 wound). Laboratory susceptibility testing revealed a high rate of resistance to ampicillin (10/16; 63%), low rate of resistance to gentamicin (1/16; 6.3%), and no resistance to ceftazidime, cefepime, or meropenem. Clones causing classic neonatal meningitis were identified (ie ST95, ST38, ST141). More recently emerged extraintestinal pathogenic (ExPEC) clones were also identified (ie ST69, ST131, ST1193), constituting 83% of isolates from infants < 27 weeks’ gestation. Numerous antibiotic resistance genes were identified in 10/16 isolates (Table and Figure), and every isolate contained multiple genes associated with disease virulence (Table).
Conclusion(s): The identified clonal diversity likely reflects both microbial evolution and changes in host populations, including evolving limits of viability and improving survival among extreme preterms. Resistance to first line antibiotics was evident from susceptibility testing and numerous antibiotic-resistance genes were identified. Large-scale genotypic and phenotypic surveillance of neonatal E. coli is warranted to inform antibiotic decisions and to identify novel strategies for prevention and early detection.
.png)