Neonatology
Session: Neonatal Neurology 10: Neurodevelopment
356 - Comparative Analysis of Rare CNVs in Chinese Neurodevelopmental Disorder Patients based on Exome Sequencing Data
Monday, May 6, 2024
9:30 AM - 11:30 AM ET
Poster Number: 356
Publication Number: 356.2918
Publication Number: 356.2918

Lin Yang, Ph.D (she/her/hers)
Shanghai
Children's Hospital of Fudan University
Shanghai, Shanghai, China (People's Republic)
Presenting Author(s)
Background: Neurodevelopmental disorders (NDDs) are a group of early-onset childhood disorders secondary to brain development and/or dysfunction. Although a large number of NDD-associated copy number variations (CNVs) have been identified using array comparative genomic hybridization techniques, there has been limited exploration of the characteristics of CNVs that confer the highest NDD risk and which subtypes carry the greatest CNV burden, particularly in non-Caucasian pediatric cohorts.
Objective: To investigate the factors responsible for conferring NDD risk via CNVs, as well as to identify which subtypes of NDD bearing the heaviest CNV burden in genome-wide scale among a large non-Caucasian cohort. Concurrently, investigate high-risk de novo CNV regions in NDD-affected children through trio family data.
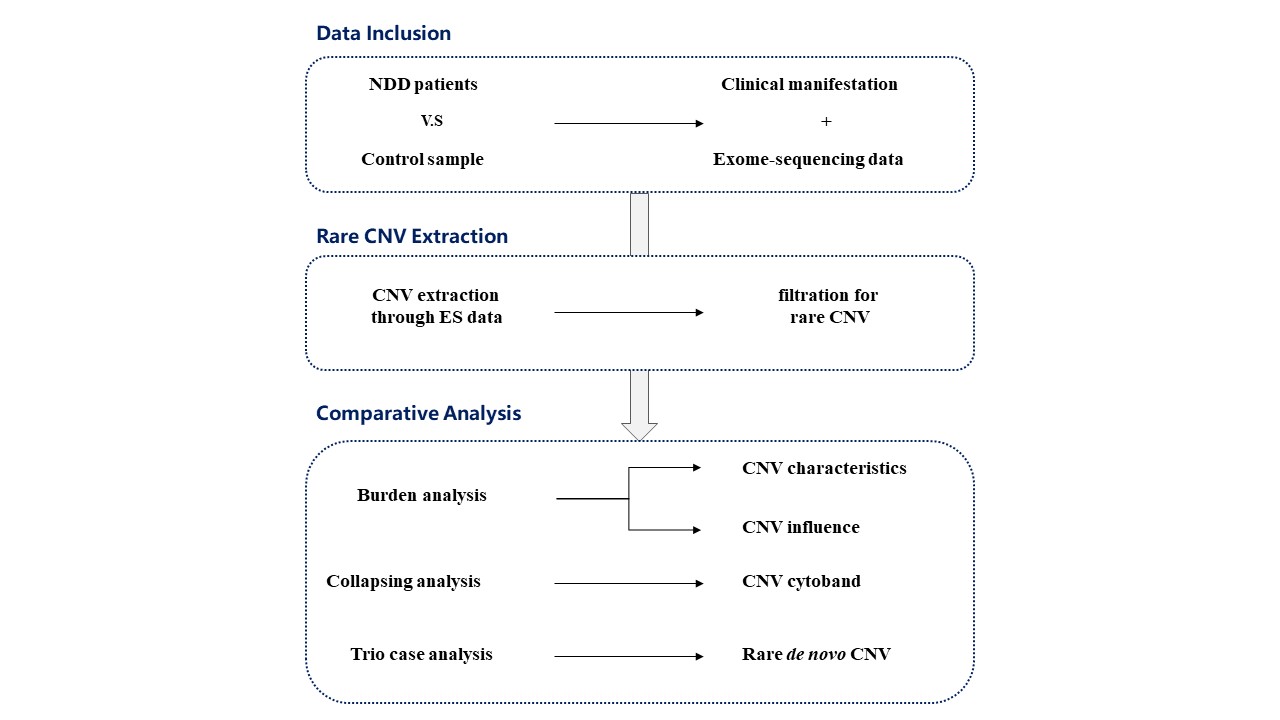
Design/Methods: In this study, we enrolled the largest NDD children and normal control cohort in China to date (n = 10,616) with clinical-exome sequencing (CES) and whole-exome sequencing (WES) data. Rare CNVs within the genomic range were detected based on sequencing data, and multi-dimensional comparative analysis was conducted to determine NDD risk factors. Additionally, we performed comparative analysis of rare de novo CNVs and analyzed related genes and biological functions using trio data.
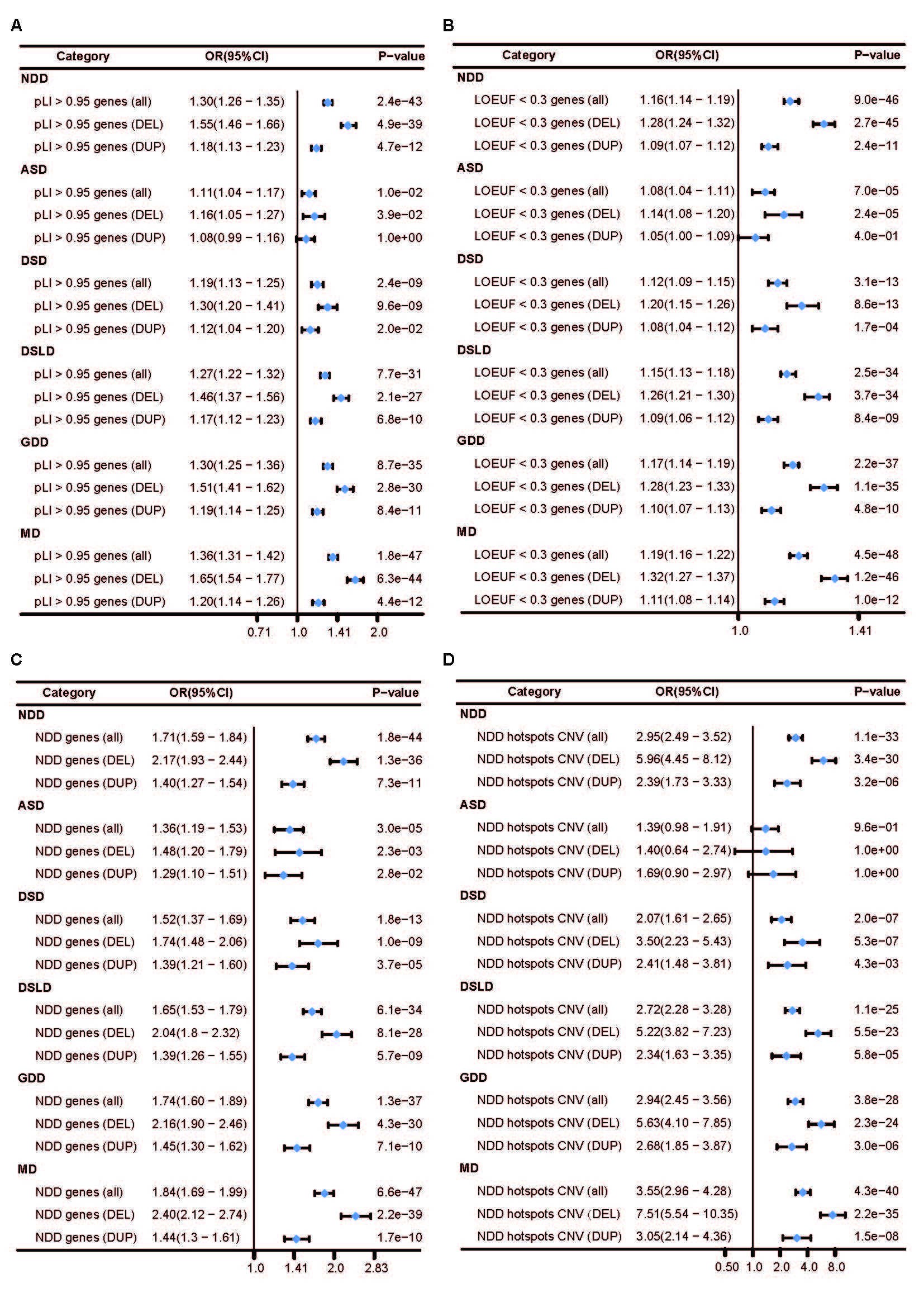
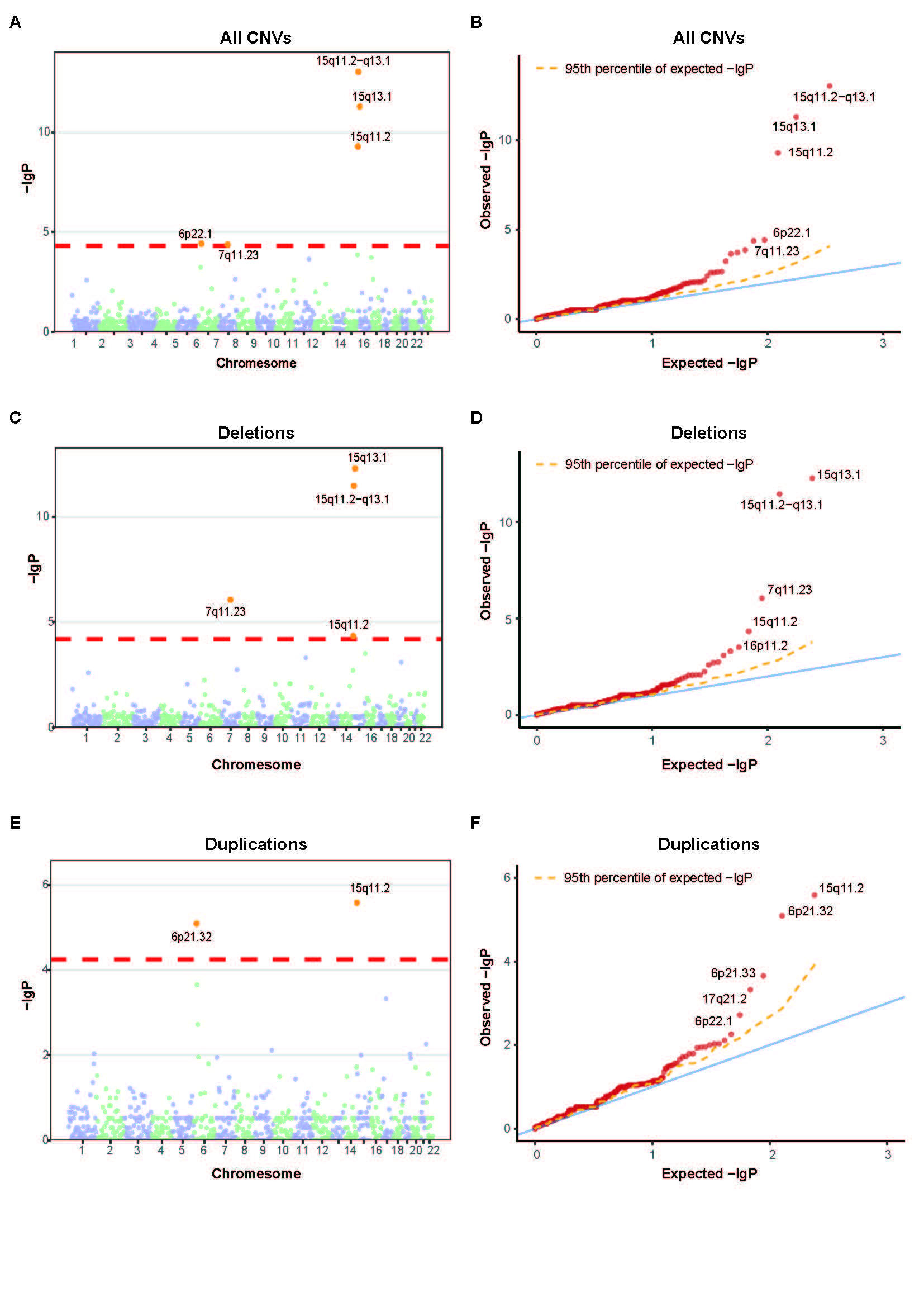
Results: Through comparative analysis of rare CNVs, this study found that in NDD and related specific subtypes of diseases, rare exonic CNVs exhibited an increased burden in the entire genome compared to normal controls. This increased burden included both the characteristics of the rare CNVs themselves and their impact on relevant genes and pathogenicity. Two novel CNV regions were identified to be potentially associated with NDD in the Chinese population, including copy number deletion at 15q13.1 and copy number duplication at 6p21.32. By performing comparative analysis of rare de novo CNVs in NDD patients and control samples with trio sequencing data, 45 genes were found to be significantly enriched in NDD patients. Biological functional analysis of the enriched genes revealed a strong association between CNVs in the GABA receptor subunit genes B3, A5, and G3 located on chromosome 15q11-q13 and the occurrence of NDD.
Conclusion(s): This study provided a comprehensive description of rare CNV burdens in the largest cohort of NDD patients and control samples in China, identifying novel CNV chromosomal regions potentially associated with pathogenicity, as well as enriched genes and biological pathways related to NDD in the Chinese population, as revealed by whole-exome sequencing data.