Nephrology
Session: Nephrology 2
22 - Methylation signatures of plasma and urine cfDNA inform host mechanisms in kidney allograft rejection and the resulting allograft response
Saturday, May 4, 2024
3:30 PM - 6:00 PM ET
Poster Number: 22
Publication Number: 22.1661
Publication Number: 22.1661

Benjamin L. Spector, MD, MS (he/him/his)
Assistant Professor of Pediatrics
University of Wisconsin School of Medicine and Public Health
Madison, Wisconsin, United States
Presenting Author(s)
Background: Acute allograft rejection (AAR) is a risk for kidney allograft failure. Knowledge of the molecular pathophysiology of AAR is lacking. Elucidating AAR immune mechanisms and the resultant allograft responses are critical to preserving allograft health. Cell-free DNA (cfDNA) is a realtime biomarker of organ pathology and cell activity. DNA methylation is a determinant of gene expression. Our prior investigations of plasma cfDNA methylation in kidney transplant recipients implicated host lipid metabolism, stress responses, and immune responses in AAR pathogenesis.
Objective: We propose urine cfDNA is derived from renal cell turnover. As such, urine cfDNA and its methylation patterns reflect the kidney response to the local environment. We aim to define allograft response to rejection by characterizing the methylomic landscape of urine cfDNA in the presence vs absence of AAR. We hope this will lay the groundwork for our long-term goal of following these methylation signatures longitudinally to identify intervenable mechanisms contributing to allograft nephropathy.
Design/Methods: DNA methylation statuses of total urine and plasma cfDNA samples from pediatric kidney transplant recipients at time of allograft biopsy were assessed using whole genome bisulfite sequencing. Differentially methylated CpG sites (DMCs) and associated differentially methylated genes (DMGs) in patients with vs without AAR were identified with logistic regression analyses and defined as ≥20% methylation difference and q-value < 0.05. DMG lists were used to identify enriched cell types contributing to urine and plasma cfDNA pools. Gene ontology (GO) analysis was performed to determine enriched biologic pathways in the urine of those with vs without AAR.
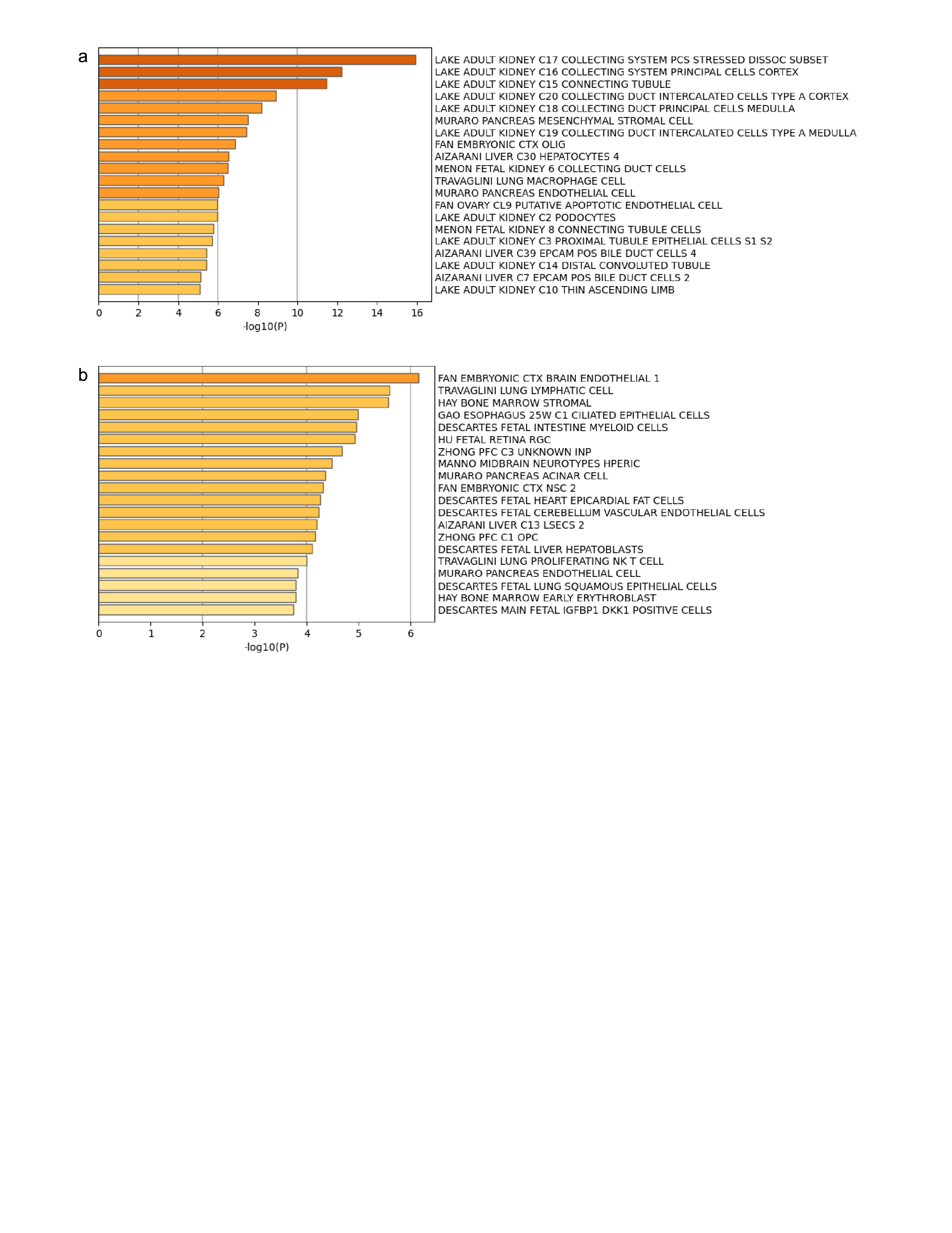
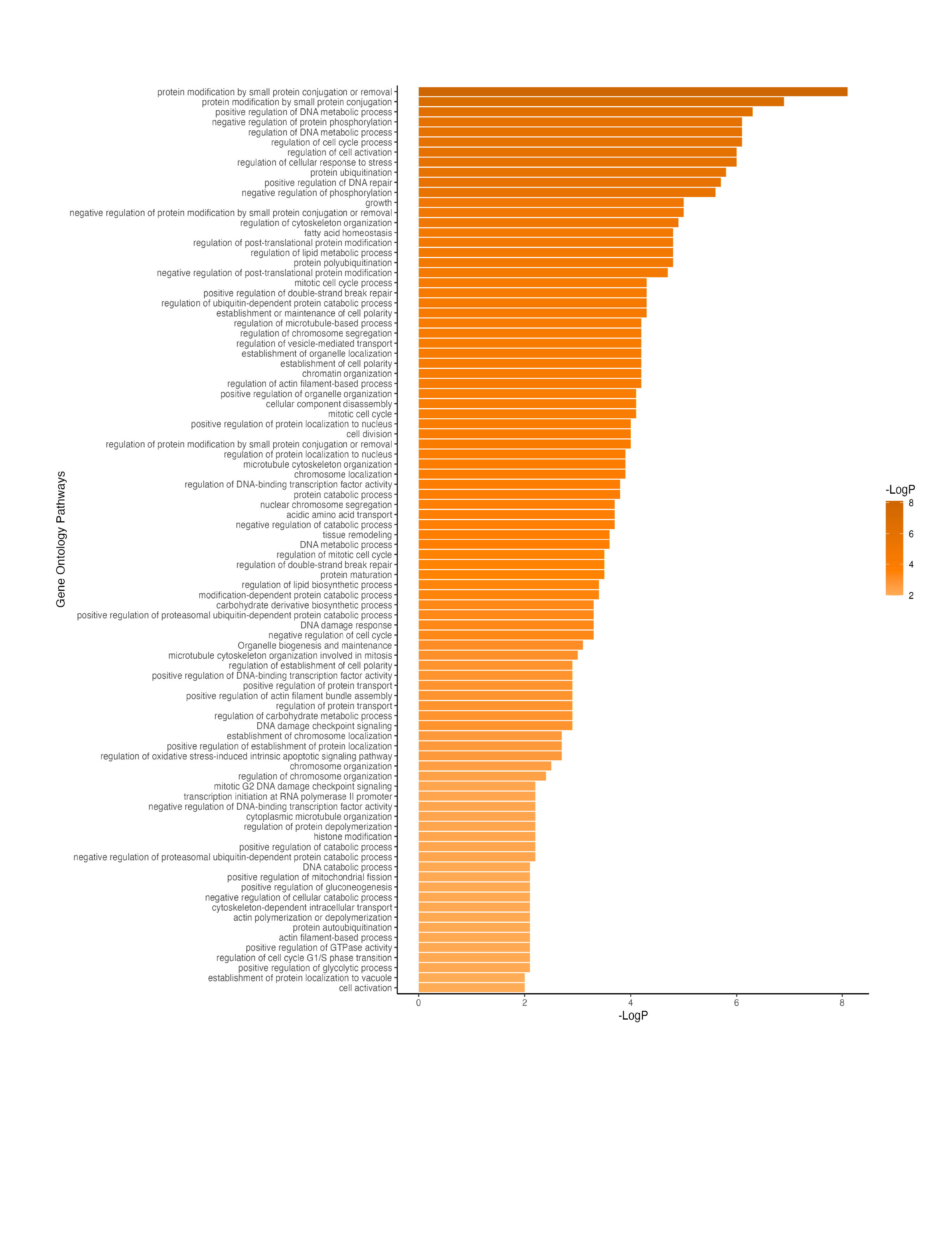
Results: In urine cfDNA we identified 9,486 DMCs, representing 595 DMGs, comparing patients with vs without AAR. Urinary DMGs were highly associated with kidney-specific cell types (Figure 1a). Plasma DMGs revealed that multiple cell types comprised the plasma cfDNA pool, with notable contributions from immune cells, erythroid precursors, and endothelial cells (Figure 1b). Enriched functional pathways associated with urinary DMGs included processes implicated in cell synthesis, DNA repair, and regulation of gene expression (Figure 2).
Conclusion(s): Our data demonstrate that kidney cells are the predominate contributors to urinary cfDNA. As such, urine cfDNA methylation can be used to inform local allograft responses to AAR, as compared to plasma cfDNA which reflects the immune response. These data suggest kidney cells undergo alterations in cell synthesis and repair in response to AAR.