Neonatology
Session: Neonatal Infectious Diseases/Immunology 3
625 - Placental Inflammation and Association with Umbilical Cord Bacteria Identified via Sequencing in Preterm Infants <1500g
Friday, May 3, 2024
5:15 PM - 7:15 PM ET
Poster Number: 625
Publication Number: 625.282
Publication Number: 625.282

Amy Hammen, DO (she/her/hers)
Clinical Fellow
Washington University in St. Louis School of Medicine
Saint Louis, Missouri, United States
Presenting Author(s)
Background: Placental inflammation is common in preterm births (up to 70%). However, the majority of cases with placental inflammation do not have an identified pathogen using culture-based methods. We hypothesize that placental inflammation is driven by infectious pathogens in utero in a high proportion of preterm infants. Umbilical cord tissue (UCT) is the ideal tissue for in utero pathogen detection as it contains fetal blood and is exposed to, colonized by, and in some cases invaded by pathogens in the amniotic fluid. Using advanced molecular assays, we may better identify presence of infectious pathogens in placentas of preterm infants exposed to in utero inflammation.
Objective: To determine whether acute placental inflammation is associated with bacteria detected in UCT by 16S rRNA gene amplicon sequencing.
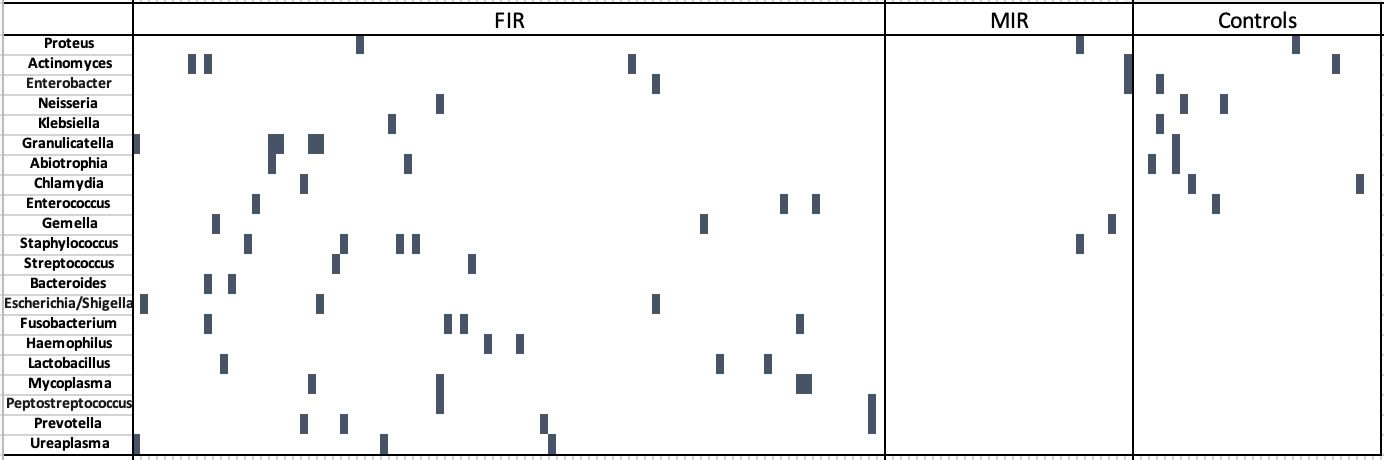
Design/Methods: We used a retrospective cohort of infants born < 1500g at a suburban Chicago hospital between 2017-2022. Acute placental inflammation (AI) was identified using Amsterdam criteria, which included staging of acute fetal inflammatory response (FIR) and maternal inflammatory response (MIR). Cases with visible bacteria on histology were identified. DNA was extracted from formalin-fixed paraffin embedded UCT and analyzed via 16S RNA gene amplicon sequencing. Identified sequences were classified to the genus level with Kraken2. Associations between genera of 21 known placental pathogens assessed by sequencing and placental inflammation were identified with Fisher’s Exact tests.
Results: Three groups were compared: FIR (n=93), MIR only (n=31), and no acute inflammation (n=32). We detected bacteria in 44% of placentas with FIR, 6% with MIR only, and 33% with no acute inflammation. Eleven bacteria were only found in inflamed cases and more frequently in those with FIR (29% of FIR and 6% of MIR P< 0.001). 16 samples with visible bacteria on histology were identified prior to sequencing, all within the group with FIR. In 12/16 samples, sequencing identified pathogenic bacterial species, none of which were detected by neonatal blood culture.
Conclusion(s): We show that sequencing techniques can identify bacteria that were missed by traditional culture methods. Detection of bacterial DNA was highest in the umbilical cords which showed FIR; however, the prevalence was not significantly different from the overall detection in uninflamed umbilical cords. Further exploration of the types of bacteria detected and thresholds for detection by these sensitive methods may help identify infants at risk for development of adverse neonatal outcomes and impact treatment of placental pathogens and/or inflammation.
.png)
