Genomics/Epigenomics
Session: Genomics/Epigenomics
375 - The role of KHDC3 & small RNAs in the epigenetic inheritance of obesity & metabolic disease
Monday, May 6, 2024
9:30 AM - 11:30 AM ET
Poster Number: 375
Publication Number: 375.2871
Publication Number: 375.2871

Sean M. Cullen, MD/PHD (he/him/his)
Instructor
Weill Cornell Medicine
New York, New York, United States
Presenting Author(s)
Background: Childhood obesity is a worsening public health epidemic in the United States. Obesity is a multifactorial disease, but it exhibits strong familial inheritance. There is a large gap in understanding of how obesity is inherited, and mouse models show a significant role for sperm small RNAs.
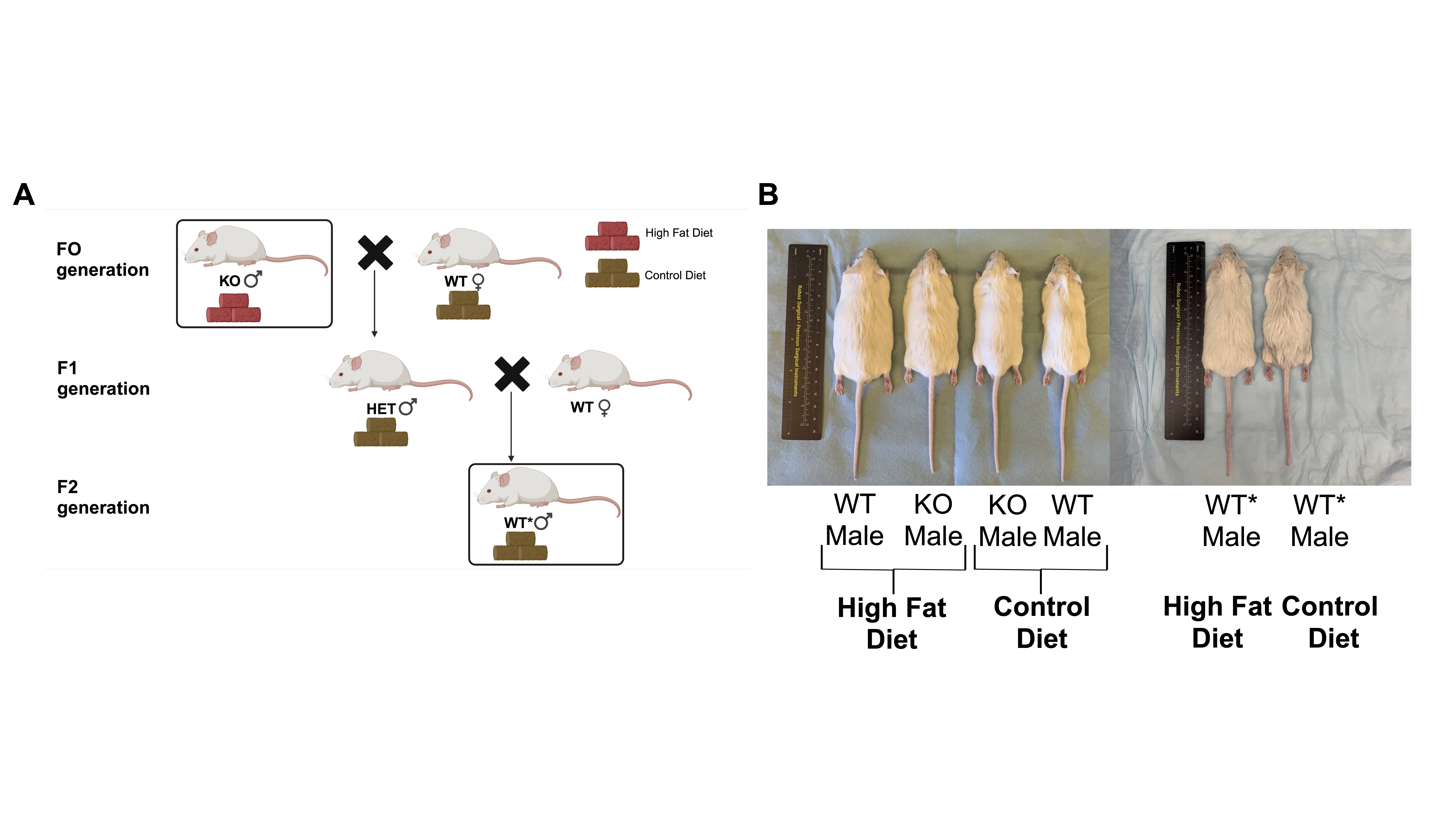
Objective: We hypothesize that KHDC3, strongly expressed in germ cells, is the key regulator for integrating external signals into changes in sperm small RNA expression patterns, which impacts the inheritance of obesity & metabolic disease. We will utilize a Khdc3-null (KO) mouse model and obesity from a high fat diet (HFD) to study these changes and determine how small RNAs lead to weight & metabolic alterations in wild-type (WT) progeny named WT* mice (Figure 1a).
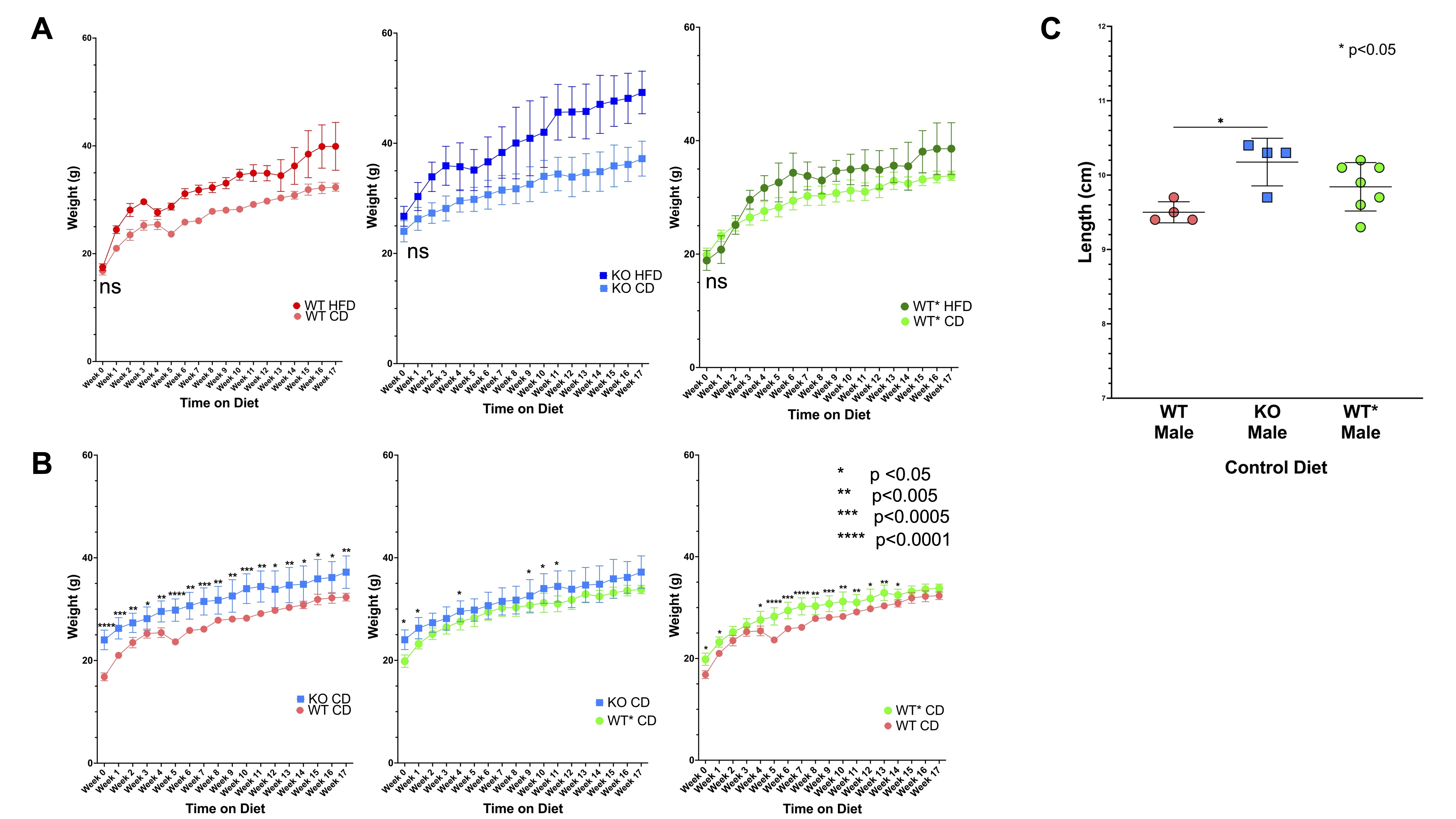
Design/Methods: KO, WT, & WT* male mice were fed a HFD or control diet (CD) for 16 weeks (Figure 1b). Males bred to WT females to generate WT progeny (WT*) were exposed to some combination of ancestral deletion of Khdc3 and/or a HFD. Males had their sperm small RNA isolated & sequenced. Weight gain was monitored weekly and glucose tolerance tests (GTTs) were administered, and tip-tail base length was noted.
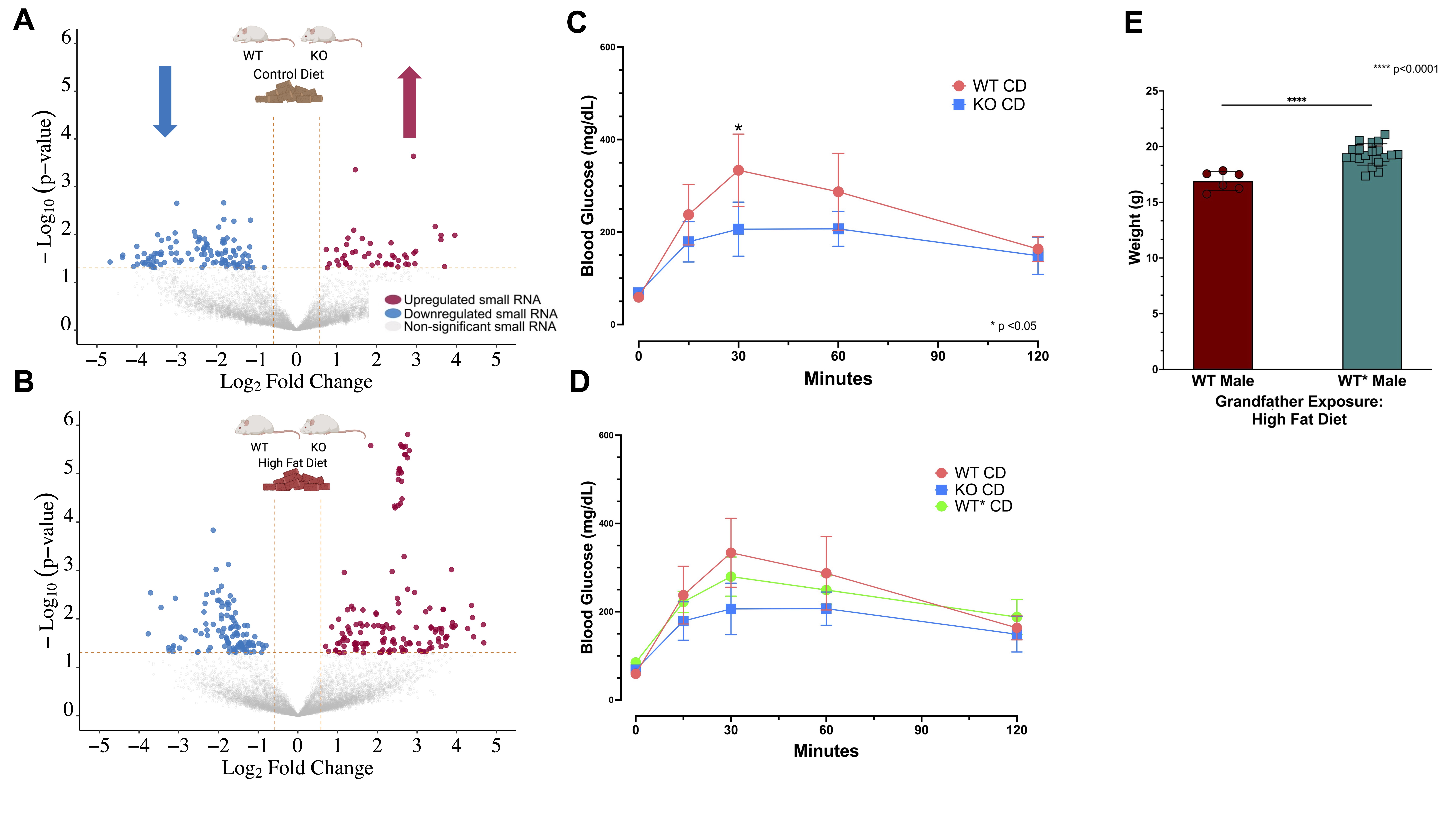
Results: All mice on a HFD gained significantly more weight than their CD counterparts (Figure 2a). KO males on CD weigh statistically more over time than WT males. WT* males do not differ statistically significantly from either WT or KO mice, an intermediate phenotype (Figure 2b). KO mice are longer than WT mice, while WT* mice are an intermediate length, but not statistically different from KO or WT males (Figure 2c). When examining the sperm small RNA components of KO & WT males exposed to either HFD or CD, there are significantly up- & down-regulated small RNAs from multiple small RNA classes, including several small RNAs involved in glucose & lipid metabolism (Figure 3a & b). KO mice fed a CD experience a blunted response to GTT, compared to WT mice (Figure 3c). There is an intermediate response to WT* mice fed a control diet (Figure 3d). Finally, WT* males at 28 days old with an ancestral exposure to both Khdc3 deletion and a HFD weigh more than WT* males alone, suggesting an additive effect (Figure 3e).
Conclusion(s): Khdc3-null mice have an observable phenotype of both larger size and improved glucose homeostasis, compared to WT mice. They also exhibit significant sperm small RNA dysregulation whether exposed to a HFD or not. There is persistence in these metabolic phenotypes in WT* males, strongly suggesting heritable changes in sperm. Future experiments will examine metabolic tissues of WT* males to determine the mechanism involved.