Technology
Session: Technology 2
196 - xBP: A Large Physiological Model-Based Generative Artificial Intelligence System That Non-Invasively Recapitulates Arterial Blood Pressure Waveforms
Sunday, May 5, 2024
3:30 PM - 6:00 PM ET
Poster Number: 196
Publication Number: 196.2167
Publication Number: 196.2167

Ben Shank, MS, Applied Mathematics
CEO
EMx Biotech LLC
Austin, Texas, United States
Presenting Author(s)
Background: The recent confluence of advanced machine learning techniques, vast amounts of high-quality data, and accessible high-performance computational capacity have led to the advent of large generative artificial intelligence (AI) models. Regarding human physiology, the integration of large physiological models into AI applications holds the promise for transformative breakthroughs in healthcare monitoring.
Objective: The goal of this study was to assess the performance of a commercial AI application in generating accurate arterial blood pressure waveforms (ABPWs) solely from electrocardiography (ECG) and photoplethysmography (PPG) data, compared to ABPWs obtained through arterial lines.
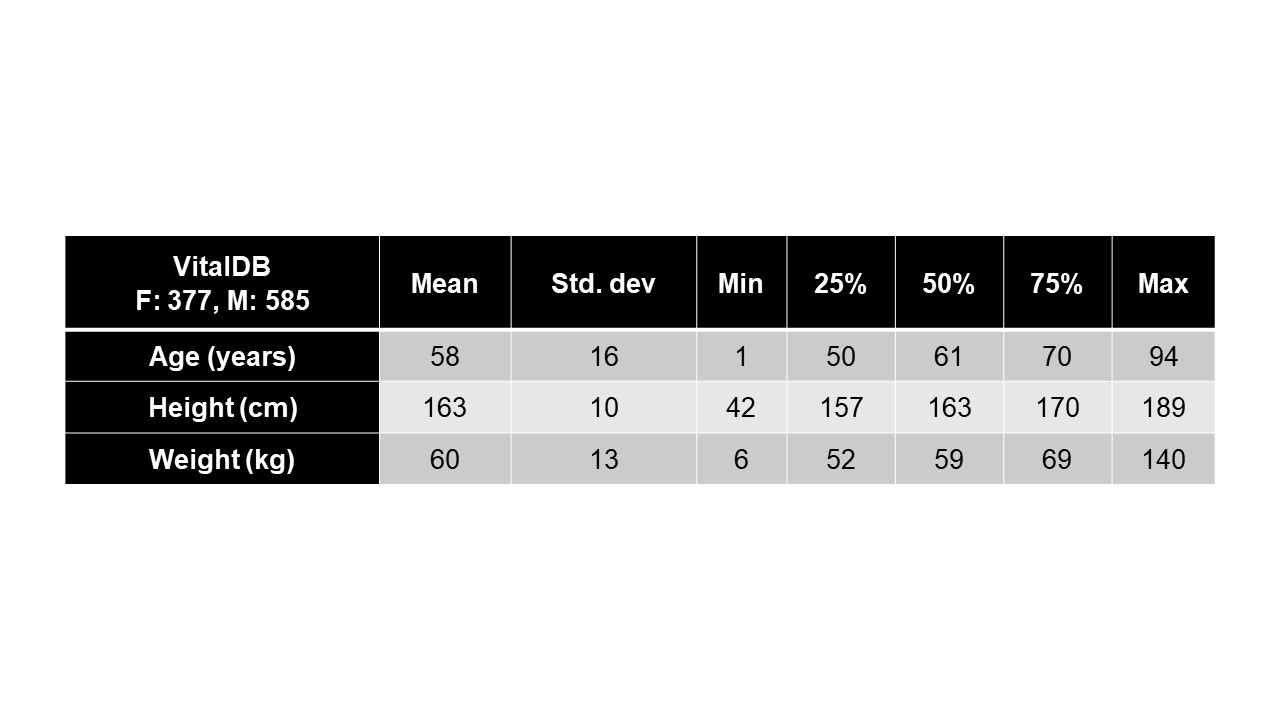
Design/Methods: EMx Biotech LLC’s xBP application is a large physiological model-based generative AI system purpose-built for invasive physiological waveform inference from non-invasive biosignal sources. The xBP base model was trained using a subset of data from VitalDB, a publicly-available repository of vital signs, consisting of approximately 2,403 hours of time-synchronized ECG, PPG, and invasive radial ABPW biosignals across 962 individual cases (see Table 1). The xBP base model was then fine-tuned using a dataset of de-identified pediatric intensive care unit patients from Texas Children’s Hospital (TCH) consisting of 89 minutes of time-synchronized ECG, PPG, and ABPWs across 19 cases. Model evaluation was performed over a TCH test set consisting of 107 minutes across 20 cases. Mean-squared error (MSE) was the only loss and evaluation metric utilized. Model training and evaluation was performed using TensorFlow 2.14.0 in Google Cloud.
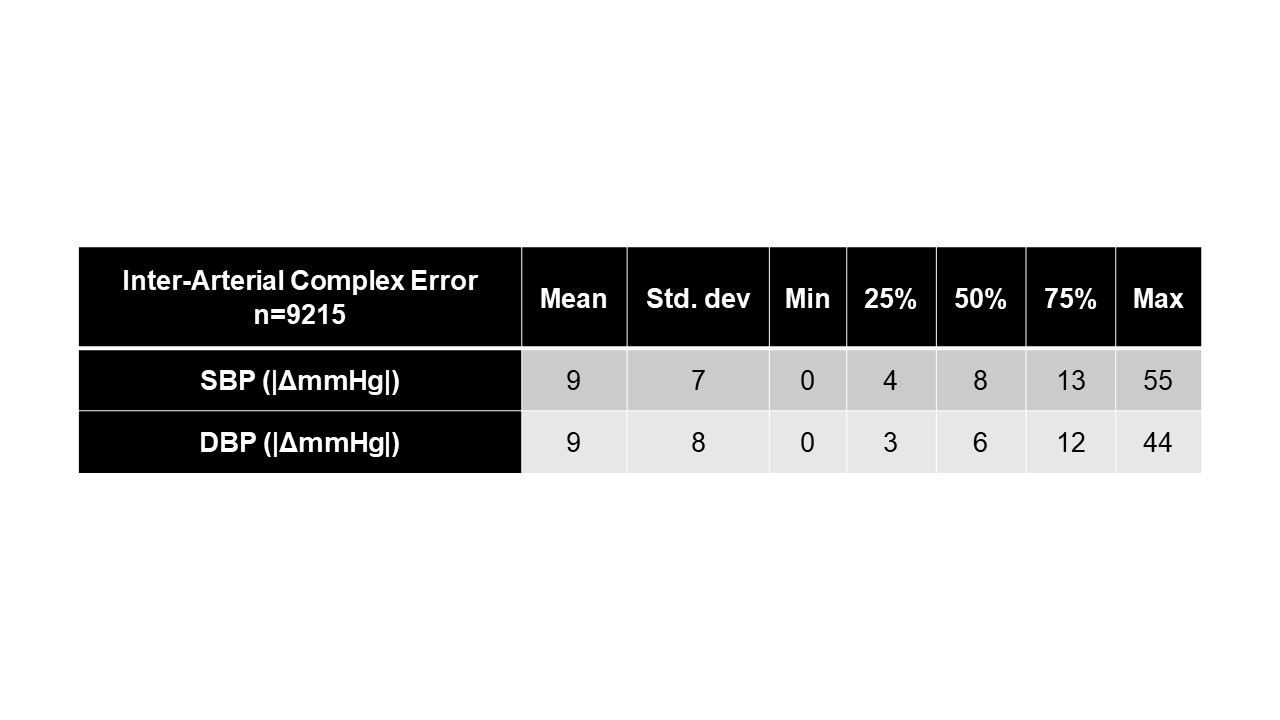
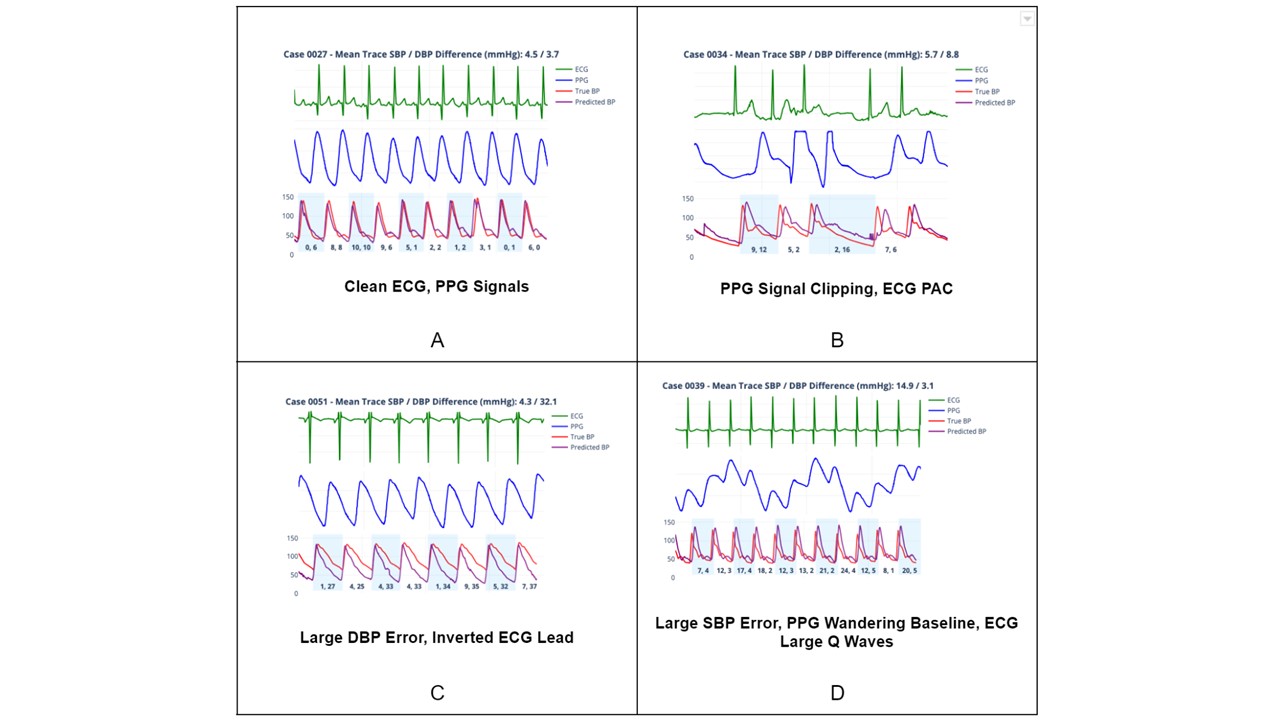
Results: Although VitalDB includes pediatric data (n=15), the median age in the training dataset was 61 years and the majority of patients were male (60.8%, n=585) with a median weight of 59 kg. The TCH test cohort included children < 18 years of age. Fine-tuning converged to minimum training and validation MSE of 4.2e-04 and 0.02, respectively. For each validation set TCH case, the absolute difference between systolic and diastolic peaks of individual true and predicted arterial complexes was calculated in mmHg. Resulting ensemble error statistics can be viewed in Table 2. Figure 1A-D illustrates the performance of xBP in four distinct scenarios.
Conclusion(s): Despite building a generative AI application in a predominantly adult population, xBP was able to generate ABPWs in children with accuracy comparable to arterial line ABPWs. Currently, we are evaluating the performance of xBP in the WAVES pediatric intensive care unit signal database (n=50,364).