Emergency Medicine
Session: Emergency Medicine 4: Infections
101 - Natural Language Processing-Assisted Evaluation of Empiric Antibiotics for Skin and Soft Tissue Infections
Saturday, May 4, 2024
3:30 PM - 6:00 PM ET
Poster Number: 101
Publication Number: 101.1631
Publication Number: 101.1631

James Rudloff, MD (he/him/his)
Clinical Fellow
Washington University in St. Louis School of Medicine
Saint Louis, Missouri, United States
Presenting Author(s)
Background: Antibiotic stewardship in all settings is a national healthcare priority, however antibiotic stewardship programs (ASPs) are limited by the need for robust data sets that often require labor-intensive chart reviews, which may be impractical.
Objective: We employed natural language processing (NLP) techniques to assess adherence to guidelines for skin and soft tissue infections (SSTIs) in a pediatric emergency department (ED).
Design/Methods: We obtained data from the electronic health record from a single urban, tertiary pediatric ED that treats approximately 50,000 patients per year. We included children aged 1-18 years based on ICD-10 codes for SSTIs; we excluded hospitalized patients. We then used Document Review Tools, a graphical-user interface, to develop an NLP model. Data analysis was performed on two datasets: a training dataset from July 1st, 2018, through June 30th, 2022, and a testing/validation dataset from July 1st, 2022, through June 30th, 2023. Using narrative documentation and prescribing information for each encounter, we evaluated whether each SSTI encounter resulted in a guideline-adherent antibiotic or a non-adherent empiric antibiotic based on the Infectious Diseases Society of America (IDSA) Guidelines. We compared the model’s performance to expert review to determine the model’s accuracy.
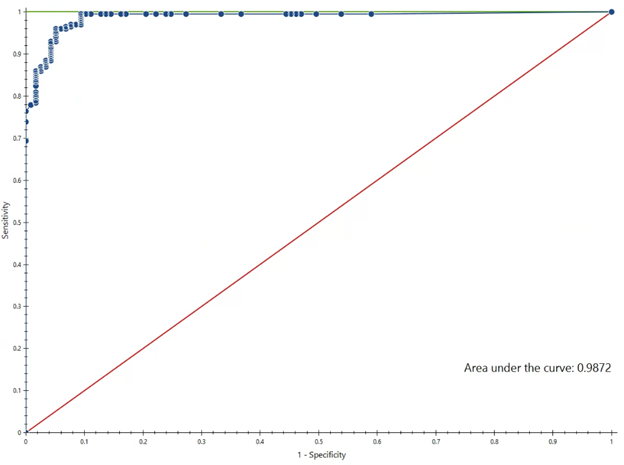
Results: The training dataset included 1588 patients, and the testing/validation dataset had 423 patients. The Random Forest (RF) model had an area under the receiver operating characteristic (ROC) curve of 0.99, indicating high accuracy in determining guideline-adherent antibiotic prescriptions (Figure 1). This training dataset achieved a sensitivity of 96.0% and a specificity of 94.9%. When applied to the testing/validation dataset, the RF model achieved a sensitivity of 96.6% and a specificity of 90.1%. When applied to all patient notes in the training dataset, the model demonstrated that 66.0% of antibiotic decisions were guideline-adherent.
Conclusion(s): We found a significant gap in guideline adherent antibiotic prescriptions for SSTIs, with about one-third of prescriptions non-adherent to the IDSA guidelines. Our novel NLP model demonstrated considerable accuracy and shows promise in supporting antibiotic stewardship programs by potentially replacing time-consuming chart reviews. Our study was limited to a single-center study and may require retraining for application in a different setting. Next steps include prospective audit and feedback locally and development of a multicenter model.