Neonatology
Session: Neonatal GI Physiology & NEC 1: Necrotizing Enterocolitis
509 - Dysbiosis and Systemic Inflammation in Necrotizing Enterocolitis: A multi-omics investigation
Sunday, May 5, 2024
3:30 PM - 6:00 PM ET
Poster Number: 509
Publication Number: 509.2058
Publication Number: 509.2058

Bryce Jorgensen, DO (he/him/his)
Neonatology Fellow
Baylor College of Medicine
Houston, Texas, United States
Presenting Author(s)
Background: Necrotizing enterocolitis (NEC) is a devastating disease of preterm infants with an incidence of 5-10% and an estimated mortality up to 30% in very low birth weight infants (VLBW). In preterm infants, NEC may be associated with dysbiosis and dysregulated inflammation.
Objective: We hypothesize that dysbiosis in the preterm intestine result in excessive inflammatory responses that contribute to NEC. Through the comparison of NEC cases to controls in this VLBW cohort we aimed to complete a multi-omics evaluation.
Design/Methods: Using a nested case-control study, a cohort of preterm infants ( < 32 weeks gestation or born at < 1500 g) were enrolled after informed consent. We will identify NEC (stage II per Bell Staging) cases and match by GA and sex with VLBW infants without NEC (controls). Stool, urine, and blood samples were collected weekly for 4 weeks. Whole genome sequencing (and microbiome) were performed in the stool samples. Microbiome Sequencing Data was analyzed through ATIMA (Agile Toolkit for Incisive Microbial Analyses) developed by CMMR of Baylor College of Medicine (BCM). Serum was assessed for systemic inflammation using a Luminex kit (for 48 cytokines and chemokines) at the proteomics core at BCM. Cytokines and Chemokines were analyzed via mixed effects linear model fit for each biomarker to compare levels in NEC patients versus controls. Urine was evaluated via by metabolomics core at BCM. T -test was utilized to analyze Urine metabolites between NEC and Control Groups.
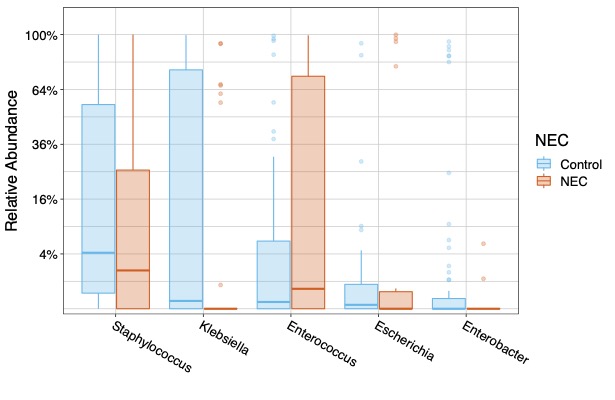
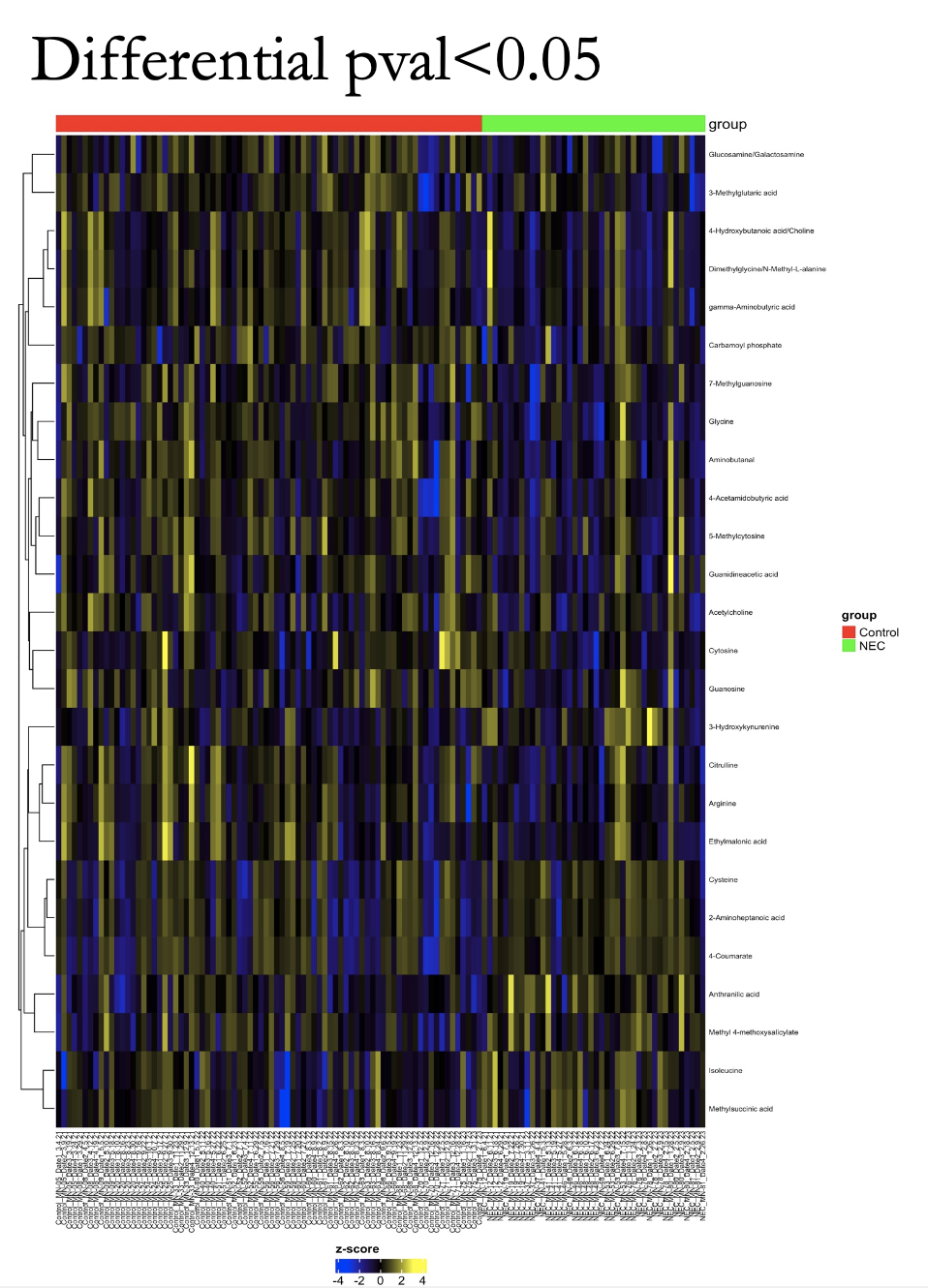
Results: We enrolled 11 NEC cases and matched 22 controls (1:2 ratio) during the study period at Texas Children’s Hospital. Microbiome analysis showed statistically significant Alpha diversity (P=0.005) and Beta Diversity (P=0.003) between the microbiome of NEC and control cases (Figure 1). Additionally, the relative abundance of genus level showed statistically significant difference in Klebsiella (P= 0.009) and Enterobacter (P=0.0141) in control cases (Figure 2). Of the 48 inflammatory markers analyzed, serum concentrations of IL 7 (P=0.02), IL 10 (P=0.03), and IL 27 (P=0.04) were found to be significantly higher in the NEC cases. Finally, distinct differences of Urine Metabolites were identified in the NEC cases (Figure 3).
Conclusion(s): We found a significant difference in the gut microbiome diversity of NEC group as well as elevated serum inflammatory markers. Urine Metabolite patterns specific to NEC cohort may be used as potential biomarkers. We speculate that the microbiome dysbiosis contributes to dysregulated inflammation seen in NEC, thus making the microbiome a potential therapeutic target.
.jpg)